
Laboratory Investigations in Microbiology

 |
Laboratory Investigations in Microbiology |
 |
Bacteriophages are viruses that can infect only bacteria. And just as animal viruses are specific to a particular host (human, dog, sheep), most bacteriophages are specific to one or a few closely related strains of bacteria. Some of the better-understood bacteriophages are lambda, T-2, and T-4 bacteriophages. Bacteriophages that infect coliform bacteria are known as coliphages (a virus that infects coliform bacteria, primarily E. coli). Phages such as T-4 are complex, naked, dsDNA viruses that attach to the bacterial cell surface and inject their DNA through the cell wall and membrane into the bacterial cytoplasm. Once inside, phage DNA is replicated and transcribed/translated by the host bacterium, producing many viral proteins and DNA which self-assemble into viral particles (virions). Eventually, the phage lyses its host cell, resulting in cell death and release of thousands of phage particles into the medium.
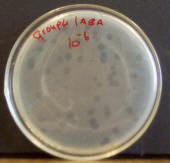 When culturing
bacteriophages in the lab, a lawn of E. coli sensitive to them must first
be grown. Virions are mixed into the agar along with E. coli cells. As the E.
coli cells start to divide and grow in the agar, bacteriophage particles attach to them,
starting the lytic life cycle. As E. coli cells nearby get infected and lysed as
well, a clear area soon begins to form around the point where the virion first
infected an E. coli cell. These clear areas are called plaques. Each
plaque was started by a single virion, and therefore the number of plaques on an
agar plate give you an indication of the number of viral particles in the
sample. (The number of plaques are used to determine the number of PFUs
(Plaque Forming Units) in a sample. A PFU is a virion that actually caused
formation of a plaque.) This method for determining number of viruses in a sample is very similar to
the viable plate count method for determining bacterial numbers, as discussed in
the previous chapter.
When culturing
bacteriophages in the lab, a lawn of E. coli sensitive to them must first
be grown. Virions are mixed into the agar along with E. coli cells. As the E.
coli cells start to divide and grow in the agar, bacteriophage particles attach to them,
starting the lytic life cycle. As E. coli cells nearby get infected and lysed as
well, a clear area soon begins to form around the point where the virion first
infected an E. coli cell. These clear areas are called plaques. Each
plaque was started by a single virion, and therefore the number of plaques on an
agar plate give you an indication of the number of viral particles in the
sample. (The number of plaques are used to determine the number of PFUs
(Plaque Forming Units) in a sample. A PFU is a virion that actually caused
formation of a plaque.) This method for determining number of viruses in a sample is very similar to
the viable plate count method for determining bacterial numbers, as discussed in
the previous chapter.
Common sources of bacteriophages include environments where such bacteria thrive, including water1 soil2, and sewage3. Bacteriophages can be isolated from these habitats by filtering or centrifuging samples to remove particulate matter (sewage, soil) and bacteria (using 0.2 micron filters). Clarified samples can then be analyzed for bacteriphages by culturing them with the appropriate host strain. Each bacteriophage has its own specific host.
In this lab exercise, we will be determining the phage titer (# phage particles/ml) of a stock preparation of T-2 Bacteriophage, as well as examining the host range of this bacteriophage.
Step 1: Serial dilution
5. Evaluate your alternate host strain plate for plaques.
References
ę 2003 - 2025 JosÚ de Ondarza, Ph.D.