
Laboratory Investigations in Microbiology

 |
Laboratory Investigations in Microbiology |
 |
Why would you want to identify an unknown bacterium? In what circumstances does this occur in real life? If you pause for a moment to think about the many ways in which microbes impact our lives, it should become clear that there are times when it is vital that we know exactly what bacteria we are facing. For example, Medical Technologists try to identify bacteria on a daily basis - bacteria which have been collected from a site of infection. Knowing which bacteria have invaded the human body is important for two reasons: 1) Diagnosis of the disease, and 2) Selecting the appropriate treatment. However, identification of 'unknown' bacteria is not limited to medicine. Researchers are constantly seeking ways to better classify (group) bacteria together based on shared characteristics, in the hope that, through understanding one bacterium, we can learn a lot about those bacteria that are closely related. Researchers are also constantly finding completely 'new' bacteria that have never been described before. Such bacteria hold the promise of new miracle drugs, novel ways for cleaning up the environment, or modern biotechnology tools.
Identification of bacteria traditionally follows a step-by-step process of
elimination. When certain characteristics are observed, we narrow down the
number of possibilities. The first step is usually a Gram
stain, which (if done
correctly) will divide bacteria up by shape, Gram reaction, and perhaps even
arrangement (strepto, diplo...). Knowing this, the scientist will carefully
choose the next step in the identification process - one that is appropriate for
the bacteria he/she has observed. Take a look at your identification chart for a
moment, and suppose your Gram stain reveals Gram-positive cocci. Immediately
your choices are narrowed down to just 11 bacteria. How would you go about
distinguishing these 11 Gram-positive cocci? Perhaps a citrate test? A catalase
test? A test of motility? An examination of the chart handed out to you in lab
will show that a citrate test is a poor choice...all 11 bacteria react the same
way, and this test does not tell you anything you did not already know!
Likewise, a test of motility is a poor choice if only two bacteria show motility
and the othe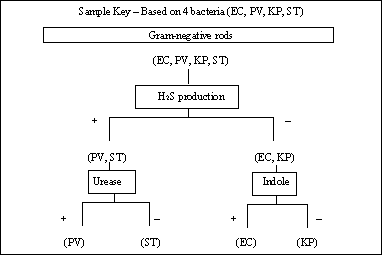 rs do not.
[Unless, of course, you have good reason to suspect one of these two bacteria to
be your unknown, based on things such as colony color]. The catalase text,
however, is an good choice - 4 cocci have catalase and 7 do not. Once you
have divided these Gram-positive cocci into these two groups, each group is
examined separately to determine the next logical test to distinguish them. For
the 4 catalase-negative cocci (EF, SL, SP, SV), perhaps a methyl red test would
work. Look at the chart and you will notice that EF and SL are MR-positive while
SP and SV are MR-negative. As a last step, one needs to now only distinguish
between two bacteria! This type of strategy described here is called a
dichotomous key strategy.
rs do not.
[Unless, of course, you have good reason to suspect one of these two bacteria to
be your unknown, based on things such as colony color]. The catalase text,
however, is an good choice - 4 cocci have catalase and 7 do not. Once you
have divided these Gram-positive cocci into these two groups, each group is
examined separately to determine the next logical test to distinguish them. For
the 4 catalase-negative cocci (EF, SL, SP, SV), perhaps a methyl red test would
work. Look at the chart and you will notice that EF and SL are MR-positive while
SP and SV are MR-negative. As a last step, one needs to now only distinguish
between two bacteria! This type of strategy described here is called a
dichotomous key strategy.
Your unknown ID project will take place over the next 2 weeks. Four days are dedicated for you to work on the unknown ID, in which time you will be expected to carry out all your identification tests. If you plan ahead, you can do one or two identification tests every lab period. You may need to come in to check on your unknown outside of regular lab periods.
Your Unknown ID will begin with you receiving broths containing your unknowns - one Gram-positive and one Gram-negative. The bacteria given to you are among those listed on the handout chart. Your first objective will be to make a dichotomous key. You should make your key to fit all the bacteria on your chart. This key is your identification strategy and you will be expected to follow it.
Your next step will be to isolate pure cultures of your two unknowns, and to perform a Gram stain on your unknowns. By streaking bacteria out on an agar plate, you will accomplish two things: 1 - separate your unknowns so you can prepare pure cultures, and 2 - observe colonies for characteristics useful in identification. Your Gram stain will eliminate many of the bacteria on the chart and is an important feature of this exercise. You can repeat your Gram stain as often as you like. Once you have pure cultures of your two unknowns, select each test you wish to perform according to your dichotomous key. Every subsequent test, of course, depends on the outcome of the previous test. In the example key shown above, the urease test would only be needed if the H2S test turned out positive! As you "order" each test, you will receive the culture media and/or reagents from me and pay $10 MicroBucks. Write your name and the name of the test you are buying on the back of the $10. Turn your inoculated media in "to be incubated" and don't forget to check on your media in a timely manner.
TSB broths containing your two unknowns
1 TSA plate
Unknown ID chart of bacteria (Excel)
Unknown ID report form (need 2 copies)
· Gram stain: can tell you shape and Gram type.
· Colony characteristics: shape, size, color, opacity
· Flagella: use Motility deeps for best determination of motility
· Endospores: May be visible in stains. Bacteria will survive 15 minutes boiling
· Fermentation tubes: type of product (acid, gas) and type of sugar fermented
· SIM deeps can tell you H2S production and Indole production.
· MR-VP broth can be used for methyl red or Voges-Proskauer test
· Casein, DNA, starch plates, gelatin deeps: hydrolysis reactions
· Citrate slants, nitrate broths, urea broths
· Catalase and Oxidase can be determined on isolated colonies on a streak plate
· Special characteristics for each of the bacteria can be found in Bergey’s Manual. Take note of these – they may help you in your identification.
· Some bacteria can appear as short rods or as cocci
· +/– indicates that the reaction is weak, slow, or variable (may be + or –); the same applies to A/– and A(g).
16. Suggested schedule
17. Hints for testing your unknowns
18. Price list
| Medium | Cost | Reagents needed | Notes |
| Staining reagents | Free | Crystal violet, Iodine, Decolorizer, Safranin | |
| TSA plates | $5 | First 2 are free | |
| TSA slants | $5 | First 2 are free | |
| TSB | $5 | First 2 are free | |
| Oxidase | $10 | Oxidase reagent | can be done on isolated colonies |
| Catalase | $10 | hydrogen peroxide | can be done on isolated colonies |
| G/L/S combination test | $10 | no Durham tube | |
| SIM | $10 | Kovacs reagent | includes indole test |
| Citrate | $10 | ||
| Starch hydrolysis | $10 | Iodine | |
| Casein hydrolysis | $10 | ||
| DNAse | $10 | 1N hydrochloric acid | |
| Gelatinase | $10 | may take 1 week | |
| Glucose fermentation | $10 | includes Durham tube | |
| Lactose fermentation | $10 | includes Durham tube | |
| Sucrose fermentation | $10 | includes Durham tube | |
| Methyl red | $10 | Methyl red reagent | |
| Nitrate reduction | $10 | Nitrate test reagent A & B; Zinc | |
| Motility test | $10 | with TTZ indicator | |
| Urease | $10 |
© 2003 - 2025 José de Ondarza, Ph.D.