Chapter 20: Microbial genetics I: control of gene expression
Even though bacteria such as E. coli have more than 1,000 genes, these are
not all continually active. Genes are said to be
expressed when their gene product (usually a protein) is being
produced by the cell. (Even then, this protein may be in an inactive or
active state). However, due to the high cost of synthesizing proteins, a
cell can conserve significant energy simply by shutting off unnecessary
genes. Enzymes for DNA replication, for example, are only needed during cell
division, and flagellar proteins only when cells are motile and planktonic.
In order to regulate the expression of a gene, cells utilize DNA sequences in
front of the protein-coding gene sequence (transcribed
region). These unique sequences (Fig. 1) are recognized by the enzyme
RNA polymerase. Additional DNA sequences may precede or follow the polymerase
binding site and allow for the attachment of protein factors that can enhance (transcription
factors) or block (repressor protein)
transcription.
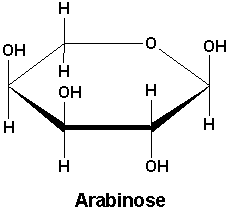 Transcription of genes in bacteria is known to be affected by oxygen,
nutrients, temperature, quorum-sensing molecules, toxins, metals, and many more.
In this experiment, we will investigate the control of the prodigiosin
pigment-producing genes in Serratia marcescens. Serratia marcescens strain 1722
(Ref. 1) contains a plasmid (pMQ262) inserted into Serratia's pigment-producing
operon, placing the pigment-producing genes under control of an arabinose-inducible
promoter. Serratia pigmentation is also temperature-sensitive.
Transcription of genes in bacteria is known to be affected by oxygen,
nutrients, temperature, quorum-sensing molecules, toxins, metals, and many more.
In this experiment, we will investigate the control of the prodigiosin
pigment-producing genes in Serratia marcescens. Serratia marcescens strain 1722
(Ref. 1) contains a plasmid (pMQ262) inserted into Serratia's pigment-producing
operon, placing the pigment-producing genes under control of an arabinose-inducible
promoter. Serratia pigmentation is also temperature-sensitive.

Materials
Per Lab group
- culture of Serratia marcescens strain 1722
- culture of Serratia marcescens wild type strain
- 1 TSA plate
- 5 TSB tubes
- 5 sterile paper discs
- arabinose and glucose solutions (10 - 100 mg/ml)
Procedure: first lab period
Arabinose
- Inoculate your TSA plate with Serratia 1722 using a cotton swab
- Aseptically place 5 paper discs on the agar plate, spaced evenly apart
- Pipette 10ul of each solution (CON, ARA 10, ARA 100, GLU 10, GLU 100) onto
a cardboard disc. Wait for solutions to be absorbed
- Place plates RIGHT-SIDE-UP into bin on the cart for incubation (48 h at
30°C)
Temperature
- Label 5 TSB tubes as 25°C, 29°C, 31°C, 33°C and 35°C
- Inoculate each test tube with 100 ul of WT Serratia culture
- Place tubes into appropriate water baths/incubators for incubation (48
h)
Procedure: Next Lab period
Arabinose
- After incubation, observe and describe your plate and the region
surrounding each paper disc
- Measure the diameter of red pigmentation around each paper disk
Temperature
Reagents: Acid-alcohol (0.02N hydrochloric acid,
95% ethanol)
- After incubation, remove tubes from water baths.
- Measure the culture density
- Pipette 1 ml of distilled water into a plastic cuvette
- Set the wavelength to 600 nm and calibrate your spectrophotometer
- ADD 0.1 ml of TSB broth from your culture tube, mix gently in the
cuvette, and record A600
- Dump the cuvette contents into the waste flask, rinse 1x with distilled
water
- Repeat for each temperature
- Extract the red pigment
- Pipette 1 ml of each TSB into a separate microcentrifuge tube; label
tubes
- Centrifuge tubes for 3 min at 13,000 rpm to pellet the cells
- Pour off the supernatant (liquid) into the WASTE flask
- Add 1 ml of acidified ethanol (wear gloves) to the pellet; mix gently by
pipetting up & down a few times
- Vortex the mixture an additional 30 - 60 seconds
- Centrifuge your tubes for 3 min at 13,000 rpm to pellet the cell debris,
leaving a clear supernatant
- Measure pigment
- Set the wavelength of a spectrophotometer to 534 nm and calibrate your
spectrophotometer using 1 ml of acidified water (aH2O)
- ADD 0.1 ml of your pigment extract to the 1 ml acidified water; mix gently
- Read the absorbance of your culture at this wavelength. Record your
data.
- Dump the cuvette contents into the waste flask, rinse 1x with distilled
water
- Repeat this procedure for each of the other 4 temperatures
- Calculate the ratio of A534/A600 for each culture.
- Plot/graph the data using Excel
References
- Khadouri, D.E. and R.M.Q. Shanks. Identification of a methicillin-resistant Staphylococcus
aureus inhibitory
compound isolated from Serratia
marcescens.
Res Microbiol. 2013 Oct; 164(8): 821–826.
- Stella, A. et al. Serratia marcescens cyclic AMP
receptor protein controls transcription of EepR, a novel regulator of
antimicrobial secondary metabolites. J. Bacteriology 197(15):2468-2478,
2015.
© 2003 - 2019 José de Ondarza, Ph.D.





Transcription of genes in bacteria is known to be affected by oxygen,
nutrients, temperature, quorum-sensing molecules, toxins, metals, and many more.
In this experiment, we will investigate the control of the prodigiosin
pigment-producing genes in Serratia marcescens. Serratia marcescens strain 1722
(Ref. 1) contains a plasmid (pMQ262) inserted into Serratia's pigment-producing
operon, placing the pigment-producing genes under control of an arabinose-inducible
promoter. Serratia pigmentation is also temperature-sensitive.
